While it is beneficial to explore novel plankton survey technology, it is essential that we also continue to maintain traditional long-term monitoring programmes to generate the necessary information to inform policy.
Please cite as: Holland, M. M., A. Atkinson, M. Best, E. Bresnan, M. Devlin, D. Johns, M. Machairopoulou, S. Pitois, J. Scott, R. Stern, C. Whyte, C. Widdicombe and A. McQuatters-Gollop (2024). Mind the Gap – The need to continue long-term plankton monitoring. Defra mNCEA Programme – Pelagic Natural Capital. Plymouth, UK, University of Plymouth. https://doi.org/10.24382/bs9p-6e92.
Changes in plankton have important implications for the continued provision of ecosystem services, including supporting commercial fish stocks, carbon sequestration, and oxygen production. Such changes can only be detected by studying long-term, consistent plankton datasets which are needed to understand the pressures driving these changes and how we can manage them.
Traditional long-term plankton monitoring relies on light microscopy to identify and count plankton taxa, with methods fully supported by national / international QA/QC standards and providing high quality trusted data.
Novel technologies, including imaging and molecular methods, offer more efficient means of collecting some types of plankton data, filling targeted knowledge gaps left by traditional monitoring. However, these data are often semi-quantitative, lacking in QA/QC standards, and/or in taxonomic resolution. While these technologies are developed it remains critical to maintain the continuity of traditional plankton monitoring to inform policy assessments of important changes in biodiversity. Losing these time-series, many of which span multiple decades, would impair our ability to detect important change in pelagic habitats, as most changes cannot be detected from short-term data. This would also accelerate the loss of taxonomic expertise, already under threat globally, diminishing our UK skill-base.
Novel technologies should be explored in parallel to traditional monitoring, as they can provide complementary data to support policy assessments and research, however, it is important that we do not attempt to replace traditional monitoring with new technology before it has been thoroughly integrated into long-term monitoring programmes.
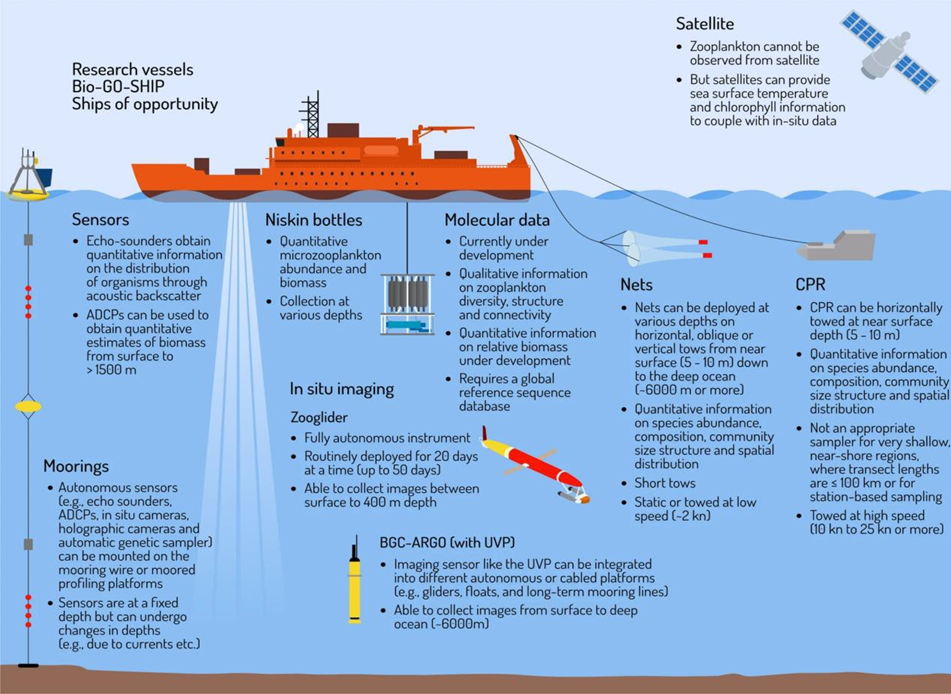
Traditional methods such as Continuous Plankton Recorder (CPR), nets and Niskin bottles have been used to monitor plankton for decades with many important research outputs. Combining traditional methods with newer methods including in situ imaging, molecular methods, advanced sensors, and satellites can improve spatial and temporal coverage and further our understanding of changes resulting from pressures on plankton communities.
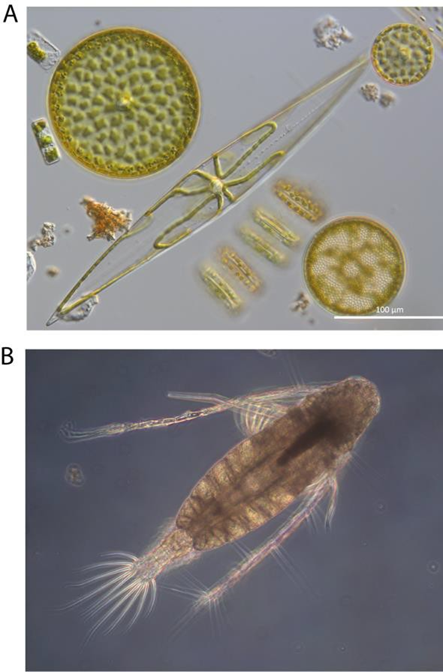
Traditional plankton monitoring involves collecting samples directly from the ocean and subsequently preserving them so they can be later analysed via light microscopy.
The CPR survey is a unique example, using a mechanical device to automatically collect and preserve samples while it is towed behind commercial ships and ferries as they travel their regular routes. The CPR is an exceptional case because it does not incur costs for research vessel time, unlike most methods, including those using novel methods.
Once on land, samples are stored and plankton entities are identified and counted in a laboratory under light microscope. Highly trained taxonomists identify and count organisms in each sample, following a consistent and documented method. Depending on the institute-specific procedure and density of organisms, sample processing time ranges from 3 hours to two days. This approach provides a high level of taxonomic detail, with semi-quantitative categories of taxa abundance. The traditional approach also allows a very rapid “sanity check” for unusual, new, and suspect taxa or results. This highly specialised method of plankton identification and enumeration is, understandably, resource intensive.
The high-quality and consistency of data collected in this manner facilitates comparisons over long time periods and between laboratories. The North-East Atlantic Marine Biological Analytical Quality Control (NMBAQC) Scheme provides a source of external Quality Assurance (QA) for laboratories engaged in the production of such marine biological data. Through the NMBAQC, laboratories engage in annual intercomparisons to ensure the phytoplankton and zooplankton data they generate are comparable to other laboratories and over time.
Traditional plankton monitoring data have revealed important large-scale declines in plankton abundance in UK waters and beyond (Edwards et al., 2022; Holland et al., 2023; Schmidt et al., 2020). Here we use the term “traditional plankton monitoring” to refer to micro-phytoplankton and zooplankton samples collected by net, bottle, bucket, or Continuous Plankton Recorder and counted by trained taxonomists using light microscopes.
Traditional plankton monitoring provides detailed abundance data which can be applied to address a wide range of questions and applications, including assessments of Good Environmental Status for OSPAR and UK Marine Strategy to inform policy decisions (McQuatters-Gollop et al., 2019; McQuatters-Gollop et al., 2022), Marine Climate Change Impacts Partnership (MCCIP) report cards to inform scientific understanding of climate change impacts on UK coasts and seas, and monitoring water quality for the Water Framework Directive (Devlin et al., 2009).
The UK’s monitoring network for plankton biodiversity includes coastal stations sampled by:
As well as and the Environment Agency (EA) for inshore water quality.
– The Scottish Environmental Protection Agency (SEPA) has stopped monitoring.
– Agri-Food and Biosciences Institute (AFBI) and Newcastle University (NU) are part of the network but have shorter time-series.
Continuous Plankton Recorder (CPR) routes displayed for visualisation, but much broader coverage is available offshore.
An excellent UK example is the Continuous Plankton Recorder (CPR) survey, the most geographically extensive marine monitoring programme in the world, with over 7 million nautical miles of tows over 90+ years, routinely counting 650+ taxa and facilitating the production of over 1000 peer-reviewed scientific publications (Richardson et al., 2006).
Similarly, Plymouth Marine Laboratory’s Western Channel Observatory L4 station is a biodiversity reference site with identification of over 500 plankton taxa alongside eDNA and benthic sampling. The sampling intensity (over 3000 net hauls since 1988) and number of variables sampled make it a testbed site for understanding how ecosystems operate, with over 350 scientific publications produced from this data (McEvoy et al., 2023).

The Marine Biological Association coordinates the Continuous Plankton Recorder (CPR) survey, employing specialised towed instruments deployed routinely from commercial ships and ferries as they travel their regular routes.
These recorders are equipped with a mechanism that allows them to filter and collect plankton samples from seawater as they are towed.
The CPR device features a silk filtering mesh that captures plankton as water passes through it, preserving a record of plankton abundance and distribution, biologically fixed in-situ.
Once in the laboratory, each roll of CPR silk is carefully unrolled, cut into sections, subsampled, and counted along transects by trained analysts with the help of light microscopes.
This innovative mechanical device has remained unchanged since it was first used in 1931.
The high frequency and broad spatial coverage of the CPR survey enable scientists to study and analyse changes in plankton communities over time, detecting important changes in pelagic habitats biodiversity and providing crucial insights into the dynamics of marine ecosystems.
Novel technologies, including automated imaging and molecular methods, are being explored as cost-effective alternatives to traditional monitoring. Automated imaging uses high-speed photography and machine learning to identify, count and measure plankton in real-time or near real-time. Molecular methods involve collection and analysis of genetic material from plankton specimens or from the pelagic environment.
While there are objectives to integrate novel technologies into routine monitoring, there are currently significant limitations to how data collected via novel technologies can be applied and a lack of long-term, consistent time-series to support assessments of pelagic habitats biodiversity. We should continue to explore these technologies, but it is essential that we do not attempt to do so at the expense of funding traditional long-term monitoring programmes, since they remain necessary to support biodiversity assessments. In addition, the taxonomic and ecological expertise to validate these novel methods needs to be maintained.
To promote their integration into routine monitoring, novel technologies should be explored in parallel to traditional monitoring to better understand how the data they generate compares to the detailed taxonomic accounting of abundance generated via traditional methods.
Limited resources and budgets for monitoring have been a major driver for technological advances made for the development of more efficient cost-effective methods to gather plankton data (Danovaro et al., 2016). Currently there is an array of novel technologies available to identify and count plankton, including molecular (Yates et al., 2019) and automated imaging methods (Pitois et al., 2018). These techniques take advantage of the latest computing and genetic sequencing technology, allowing for a far greater throughput of samples than could ever be achieved manually.
Imaging instruments, combined with machine learning to automatically classify the collected images, have received a high level of interest, due to their ability to provide rapid and unbiased data that can be stored digitally and quickly made available for use (Giering et al., 2022). Thus, they can overcome many of the limitations characteristic of traditional methods of collecting and analysing plankton samples. For example, the Plankton Imager (PI, Cefas) has been used aboard the RV Cefas Endeavour since 2016. While the ship is underway, seawater is pumped through a flow cell and a high-speed camera captures images of all passing particles. Each image is automatically classified as either copepod or non-copepod by a machine learning algorithm as well as recording size information in real-time. The system works continuously for the duration of a survey (Pitois et al., 2018; Scott et al., 2021; Scott et al., 2023).

The Plankton Imager, recently developed by Cefas and Plankton Analytics in the UK, is an imaging tool for sampling zooplankton without the need of human intervention.
It consists of a high-speed camera that images all passing particles in a flow of pumped seawater. Images are identified in real-time and uploaded via satellite. This system requires no on-board expertise, harmful chemicals, or deployment of gear from the ship.
It can collect data at a significantly finer spatial resolution than traditional methods. It also measures zooplankton, supporting carbon accounting. In its current state of development, its main capability is counting copepods.
Similarly, instruments such as the Imaging FlowCytobot (IFCB; McLane Labs) uses artificial intelligence to analyse phytoplankton images and can obtain quantitative results and accuracies comparable to human analysts. However, these methods generally only classify individual organisms into coarse groups, missing the taxonomic detail that can be achieved by human analysts, and what is often needed to understand important changes in pelagic biodiversity.
Molecular methods, such as eDNA and metabarcoding use genetic information to study and identify the various species in an ecosystem. All molecular methods require the collection of physical samples prior to analysis in a laboratory. To derive abundance (and biomass) data from eDNA, promising links have been shown between eDNA and cell biovolumes (Song & Liang, 2023). The main advantage of molecular tools is their ability to detect a broad array of taxa, including rare species, and in targeted tests provides the highest level of taxonomic detail. They can also be valuable for studying delicate organisms which can be easily damaged or destroyed by net sampling (Govindarajan et al., 2021). However, since molecular methods rely on trace amounts of genetic material there is always a risk of generating false positives, or detecting taxa which are not actually present in a sample. Additional information on sex, life stage and individual conditions are impossible to gather using eDNA approaches. Currently, eDNA techniques are also limited in their ability to estimate abundance of taxa (Yates et al., 2019) and are best suited for the provision of presence/absence data.
An important difference between traditional and emerging plankton sampling approaches is the issue of scale (Scott et al., 2023). Traditional approaches frequently integrate temporally over a week to a month and spatially, to 10 nautical miles in the case of the CPR. For the latest autonomous imaging approaches the integration scales are much finer, typically resolved to meters vertically and horizontally, and over minutes to hours for glider-, ship- or buoy- mounted instruments. This enables fundamentally different areas of science to be explored. For example, thermocline, diel cycle, or tidal cycle dynamics, impacts of extreme events such as storms or floodwater discharge or predator prey patch interactions. Obviously, however, these instruments and surveys applying them have not been running long enough to have the statistical power to resolve climate change scales. Moreover, since they span such different scales as well as sampling in a different manner, intercalibration with traditional monitoring is highly challenging. This fine level of detail is also typically not necessary for addressing policy needs.
While novel methods may appear more efficient than traditional methods, this may not necessarily be the case. There are significant hurdles involved with operationalising any novel method into practical routine monitoring. It will likely take decades before novel methods can demonstrate the consistent high-quality results of the traditional methods in use today. Currently, most imaging and molecular methods are research oriented or specialised towards a small suite of target species that are insufficient for assessing biodiversity status for policy.
Many of the UK’s traditional plankton monitoring programmes have been ongoing since the 1990s and early 2000s, with the CPR commencing much earlier in 1931. Most programmes have employed the same equipment and methods since they were initiated to maintain the comparability of a time-series, despite the emergence of improved technology and methods. Due to the highly variable and patchy distribution of plankton in both space and time, long-term monitoring is mandatory for detecting important changes. Hydrological processes, such as the Atlantic Multidecadal Oscillation (AMO) and North Atlantic Oscillation (NAO), generate pressures on plankton communities lasting several decades. Changes in plankton communities resulting from human impacts can often be overshadowed by the variability attributed to these natural cyclical processes (Harris et al., 2014). Thus, long-term contiguous data is necessary for detecting the effects of climate change, shifting circulation patterns, and rising sea levels. We also need to continuously update long-term time-series to understand ecological change since we can only detect change by comparing current conditions to previous conditions. Therefore, the value of a time-series for addressing ecological questions is inextricably linked to its consistency and duration (Brander & Drinkwater, 2003).
Research funding is increasingly allocated to exploring novel and innovative methods and technologies and it is becoming more difficult to obtain funding for routine monitoring or biodiversity assessments. In the 1980s the CPR survey was almost lost due to funding constraints. If the survey had not been saved, our current understanding of climate change and its impacts on marine biodiversity would be severely hampered (Reid et al., 2003; Richardson et al., 2006). Continued cuts to monitoring budgets have resulted in the loss of several CPR routes, temporarily reinstated through the mNCEA programme, however, future funding remains uncertain. Multiple fixed-point stations are currently in a precarious status, including the Western Channel Observatory, subject to significant funding reductions. In Scotland, SEPA have discontinued plankton monitoring, the LPO has no dedicated funding to continue, and the Scottish Coastal Observatory has experienced significant reductions in taxonomic resource, impacting the volume of samples that can be analysed.
The erosion of monitoring programme funding has also contributed to reducing taxonomic capability within the UK research community and beyond. With the current inability to recruit junior taxonomists, often driven by a lack of resources, those who remain have little time to expand their skill set to focus on emerging species of concern. Critical skills are also lost when experienced taxonomists leave or retire (see McQuatters-Gollop et al., 2017). Research roles of the future will require taxonomic skills to, for example, train Artificial Intelligence classification models, build genetic databases, and perform validation between traditional and novel methods. There needs to be a stronger recognition of the essential role that long-term time-series and taxonomic skills play in the development and incorporation of new technologies into ecological assessments. Without the traditional taxonomic skill set, these novel methods cannot be validated. Maintaining traditional long-term monitoring programmes will provide opportunities and incentives to promote training in taxonomy and will foster accelerated development of novel technology.
While imaging (Ostle & Hélaouët, 2023) and molecular (Suter et al., 2021) technologies lack the duration of use of traditional plankton monitoring time-series, the information they provide can be complementary to traditional approaches (Ratnarajah et al., 2023).
Due to their diverse range of sizes and patterns of distribution, no single method can effectively sample the full plankton community, leading researchers to select the most appropriate method to fulfil particular research or policy aims (Owens et al., 2013; Skjoldal et al., 2013). Novel technologies can provide efficient alternatives for addressing questions related to plankton distribution in space and time (Scott et al., 2021), targeted detection of harmful algal bloom species (Medlin & Orozco, 2017), and new migrants or alien species (Créach et al., 2021), however, traditional methods and associated skills remain critical to obtaining a detailed taxonomic accounting of the plankton community and validating novel methods. Most importantly, only traditional methods have decades of associated historical data required to detect long-term ecological changes.
We should be embracing novel technology, while also ensuring the continuity of traditional monitoring time-series. We cannot simply switch from traditional to novel methods since the continuity of long time-series is critical to supporting biodiversity assessments (Brander & Drinkwater, 2003). As technology continues to improve, it is possible that traditional net sampling will become less important (Giering et al., 2022), however, until this occurs we must find ways to maintain taxonomic skills and apply traditional and novel methods in a complementary manner to facilitate ongoing scientific progress.
Please cite as: Holland, M. M., A. Atkinson, M. Best, E. Bresnan, M. Devlin, D. Johns, M. Machairopoulou, S. Pitois, J. Scott, R. Stern, C. Whyte, C. Widdicombe and A. McQuatters-Gollop (2024). Mind the Gap – The need to continue long-term plankton monitoring. Defra mNCEA Programme – Pelagic Natural Capital. Plymouth, UK, University of Plymouth. https://doi.org/10.24382/bs9p-6e92.
Bedford, J., Ostle, C., Johns, D. G., Atkinson, A., Best, M., Bresnan, E., Machairopoulou, M., Graves, C. A., Devlin, M., & Milligan, A. (2020). Lifeform indicators reveal large‐scale shifts in plankton across the North‐West European shelf. Global Change Biology, 26(6), 3482-3497.
Brander, K., & Drinkwater, K. (2003). The relationship between scientific understanding and the length of time series: the CPR example. Content/Table des matières, 9.
Créach, V., Derveaux, S., Owen, K. R., Pitois, S., & Antajan, E. (2021). Use of environmental DNA in early detection of Mnemiopsis leidyi in UK coastal waters. Biological Invasions, 1-10.
Danovaro, R., Carugati, L., Berzano, M., Cahill, A. E., Carvalho, S., Chenuil, A., Corinaldesi, C., Cristina, S., David, R., & Dell’Anno, A. (2016). Implementing and innovating marine monitoring approaches for assessing marine environmental status. Frontiers in Marine Science, 3, 213.
Devlin, M., Barry, J., Painting, S., & Best, M. (2009). Extending the phytoplankton tool kit for the UK Water Framework Directive: indicators of phytoplankton community structure. Hydrobiologia, 633, 151-168.
Edwards, M., Beaugrand, G., Kléparski, L., Hélaouët, P., & Reid, P. C. (2022). Climate variability and multi-decadal diatom abundance in the Northeast Atlantic. Communications Earth & Environment, 3(1), 1-8.
Giering, S. L., Culverhouse, P. F., Johns, D. G., McQuatters-Gollop, A., & Pitois, S. G. (2022). Are plankton nets a thing of the past? An assessment of in situ imaging of zooplankton for large-scale ecosystem assessment and policy decision-making. Frontiers in Marine Science, 9, 986206.
Govindarajan, A. F., Francolini, R. D., Jech, J. M., Lavery, A. C., Llopiz, J. K., Wiebe, P. H., & Zhang, W. (2021). Exploring the use of environmental DNA (eDNA) to detect animal taxa in the mesopelagic zone. Frontiers in Ecology and Evolution, 9, 574877.
Harris, V., Edwards, M., & Olhede, S. C. (2014). Multidecadal Atlantic climate variability and its impact on marine pelagic communities. Journal of Marine Systems, 133, 55-69.
Holland, M. M., Louchart, A., Artigas, L. F., Ostle, C., Atkinson, A., Rombouts, I., Graves, C. A., Devlin, M., Heyden, B., Machairopoulou, M., Bresnan, E., Schilder, J., Jakobsen, H. H., Lloyd-Hartley, H., Tett, P., Best, M., Goberville, E., & McQuatters-Gollop, A. (2023). Major declines in NE Atlantic plankton contrast with more stable populations in the rapidly warming North Sea. Science of the total environment, 165505.
McEvoy, A. J., Atkinson, A., Airs, R. L., Brittain, R., Brown, I., Fileman, E. S., Findlay, H. S., McNeill, C. L., Ostle, C., Smyth, T. J., Somerfield, P. J., Tait, K., Tarran, G. A., Thomas, S., Widdicombe, C., Woodward, M., Beesley, A., Conway, D. V. P., Fishwick, J., . . . Widdicombe, S. (2023). The Western Channel Observatory: a century of physical, chemical and biological data compiled from pelagic and benthic habitats in the Western English Channel. Earth System Science Data, 2023, 1-42.
McQuatters-Gollop, A., Atkinson, A., Aubert, A., Bedford, J., Best, M., Bresnan, E., Cook, K., Devlin, M., Gowen, R., & Johns, D. G. (2019). Plankton lifeforms as a biodiversity indicator for regional-scale assessment of pelagic habitats for policy. Ecological Indicators, 101, 913-925.
McQuatters-Gollop, A., Guérin, L., Arroyo, N. L., Aubert, A., Artigas, L. F., Bedford, J., Corcoran, E., Dierschke, V., Elliott, S. A. M., Geelhoed, S. C. V., Gilles, A., González-Irusta, J. M., Haelters, J., Johansen, M., F, Lynam, C. P., Niquil, N., Meakins, B., Mitchell, I., . . . Vina-Herbon, C. (2022). Assessing the state of marine biodiversity in the Northeast Atlantic. Ecological Indicators, 141, 109148. https://doi.org/https://doi.org/10.1016/j.ecolind.2022.109148
McQuatters-Gollop, A., Johns, D. G., Bresnan, E., Skinner, J., Rombouts, I., Stern, R., Aubert, A., Johansen, M., Bedford, J., & Knights, A. (2017). From microscope to management: the critical value of plankton taxonomy to marine policy and biodiversity conservation. Marine Policy, 83, 1-10.
Medlin, L. K., & Orozco, J. (2017). Molecular techniques for the detection of organisms in aquatic environments, with emphasis on harmful algal bloom species. Sensors, 17(5), 1184.
Ostle, C., & Hélaouët, P. (2023). The Continuous Plankton Recorder as a platform for sensor development. PICES Press, 31(2), 64-65.
Owens, N., Hosie, G., Batten, S., Edwards, M., Johns, D., & Beaugrand, G. (2013). All plankton sampling systems underestimate abundance: response to “Continuous plankton recorder underestimates zooplankton abundance” by JW Dippner and M. Krause. Journal of Marine Systems, 128, 240-242.
Pitois, S. G., Tilbury, J., Bouch, P., Close, H., Barnett, S., & Culverhouse, P. F. (2018). Comparison of a cost-effective integrated plankton sampling and imaging instrument with traditional systems for mesozooplankton sampling in the Celtic Sea. Frontiers in Marine Science, 5, 5.
Ratnarajah, L., Abu-Alhaija, R., Atkinson, A., Batten, S., Bax, N. J., Bernard, K. S., Canonico, G., Cornils, A., Everett, J. D., & Grigoratou, M. (2023). Monitoring and modelling marine zooplankton in a changing climate. Nature Communications, 14(1), 564.
Reid, P. C., Colebrook, J., Matthews, J., Aiken, J., & Team, C. P. R. (2003). The Continuous Plankton Recorder: concepts and history, from Plankton Indicator to undulating recorders. Progress in oceanography, 58(2-4), 117-173.
Richardson, A., Walne, A., John, A., Jonas, T., Lindley, J., Sims, D., Stevens, D., & Witt, M. (2006). Using continuous plankton recorder data. Progress in oceanography, 68(1), 27-74.
Schmidt, K., Birchill, A. J., Atkinson, A., Brewin, R. J., Clark, J. R., Hickman, A. E., Johns, D. G., Lohan, M. C., Milne, A., & Pardo, S. (2020). Increasing picocyanobacteria success in shelf waters contributes to long‐term food web degradation. Global Change Biology, 26(10), 5574-5587.
Scott, J., Pitois, S., Close, H., Almeida, N., Culverhouse, P., Tilbury, J., & Malin, G. (2021). In situ automated imaging, using the Plankton Imager, captures temporal variations in mesozooplankton using the Celtic Sea as a case study. Journal of plankton research, 43(2), 300-313.
Scott, J., Pitois, S., Creach, V., Malin, G., Culverhouse, P., & Tilbury, J. (2023). Resolution changes relationships: Optimizing sampling design using small scale zooplankton data. Progress in oceanography, 210, 102946.
Skjoldal, H. R., Wiebe, P. H., Postel, L., Knutsen, T., Kaartvedt, S., & Sameoto, D. D. (2013). Intercomparison of zooplankton (net) sampling systems: Results from the ICES/GLOBEC sea-going workshop. Progress in oceanography, 108, 1-42.
Song, J., & Liang, D. (2023). Community structure of zooplankton and its response to aquatic environmental changes based on eDNA metabarcoding. Journal of hydrology, 622, 129692.
Suter, L., Polanowski, A. M., Clarke, L. J., Kitchener, J. A., & Deagle, B. E. (2021). Capturing open ocean biodiversity: comparing environmental DNA metabarcoding to the continuous plankton recorder.Molecular ecology, 30(13), 3140-3157.
Yates, M. C., Fraser, D. J., & Derry, A. M. (2019). Meta‐analysis supports further refinement of eDNA for monitoring aquatic species‐specific abundance in nature. Environmental DNA, 1(1), 5-13.